- 글번호
- 949877
First identification of microRNA (miRNA) transcription control mechanisms in response to plant environmental stress
- Writer
- 관리자
- View
- 2749
- Date
- 2023.05.30
- 수정일
- 2023.05.30
A research team led by Prof. Dae-Jin Yun of the Department of Biomedical Science & Engineering at the KU Institute of Science and Technology first identified the mechanism of transcriptional regulation of microRNA (MicroRNA, miRNA) in response to plant stress, and published the results in “Plant Cell (IF=12.08),” a renowned academic journal in the field of botany. Published online on May 18th 2023.
※ Title: The HOS15-HDA9 complex associates with HYL1 to modulate miRNA expression in response to ABA signaling
※ Lead authors: Professor Dae-Jin Yun (corresponding author, Konkuk University), Dr. Junghoon Park (first author, Konkuk University), Jane Bard (co-first author, Konkuk University)
Plants that have to live a fixed life face various environmental stresses throughout their lives, and build various defense systems to adapt to and survive in the changed environment. The micro RNA (miRNA), known as non-coding RNA transcript of 21 to 24 nucleotides, binds to complementary mRNA and cuts the mRNA or inhibits translation, thereby suppressing the expression of the target gene. The regulation of miRNA biogenesis is crucial for maintaining plant homeostasis under biotic and abiotic stress. The crosstalk between the RNA polymerase II (Pol-II) complex and the miRNA processing machinery has emerged as a central hub modulating transcription and co-transcriptional processing of primary miRNA transcripts (pri-miRNAs). However, it remains unclear how miRNA-specific transcriptional regulators recognize MIRNA loci.
Professor Yun's team discovered a protein called HIGH EXPRESSION OF OSMOTICALLY 41 RESPONSIVE GENE15 (HOS15), which is expected to play an important role in the transcriptional regulation of miRNA, through molecular genetic methods (Figure 1). It was found that they form a complex with HISTONE DEACETYLASE 9 (HDA9) and bind to HYPONASTIC LEAVES 1 (HYL1), an important protein involved in miRNA biosynthesis. In the presence of a plant stress hormone ABA, hos15/hda9 mutants show enhanced transcription of pri-miRNAs that is accompanied by increased processing, leading to over-accumulation of a set of mature miRNAs. Moreover, upon recognition of the nascent pri-miRNAs, the ABA-induced recruitment of the HOS15-HDA9 complex to MIRNA loci is guided by HYL1. The HYL1-dependent recruitment of the HOS15-HDA9 complex to MIRNA loci suppresses expression of MIRNAs and processing of pri-miRNA. Most importantly, these findings indicate that nascent pri-miRNAs serve as scaffolds for recruiting transcriptional regulators, specifically to MIRNA loci. This study proposes an intriguing model (Figure 2) for plants to regulate miRNA production under ABA treatment. The HOS15-HDA9 complex uses HYL1 and pri-miRNA as the scaffold to target the upstream of MIRNA loci, remove histone acetylation marks, and repress the miRNA processing. Considering the known functions of HYL1 as regulator of transcription and stability, this work shows that HOS15-HDA9 can regulate some MIRNA genes at both transcriptional and processing levels upon interaction with HYL1.
This work was supported by grants from the National Research Foundation of Korea funded by the Korean Government.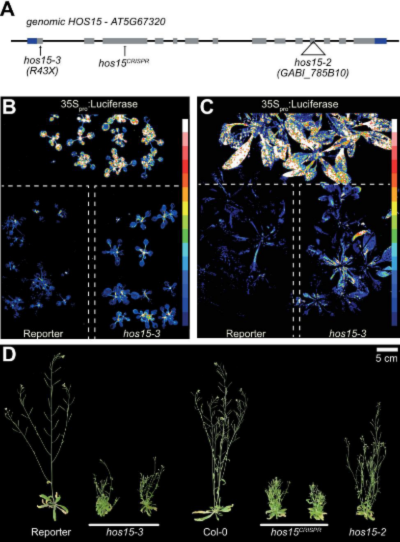
Figure 1. Identification of HOS15 involved in the transcriptional regulatory mechanism of miRNA. (A) Gene structure of genomic locus encoding HOS15 (gHOS15) showing a T-DNA insertion site in hos15-2, a nonsense mutation at amino acid residue 43; Arginine (R) in hos15-3, and the target region for the CRISPR system (hos15CRISPR). Gray boxes, exons; black lines, introns; flanking blue boxes, 5′ and 3′ UTR regions. (B and C) Bioluminescence activity as measured with a CCD camera in 14-day-old (B) and 25- day-old (C) hos15-3 mutants and control plants (reporter and 35Spro:Luciferase). The color ranging from low (blue) to high (white) represents the luminescence intensity. (D) Phenotypic characterization of control lines (reporter and Col-0), and hos15 mutants (hos15- 2, hos15-3, hos15CRISPR) having fully expanded siliques. Bar = 5 cm. To facilitate comparison and observation, each plant was photographed separately, digitally extracted, and mounted on a single panel with a black background.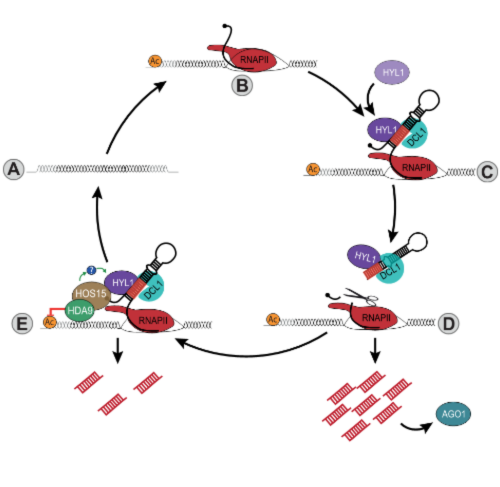
Figure 2. A model showing how transcriptionally inactive MIRNA loci (A) can be transcribed by Poll II (B) to produce a pri-miRNA transcript that folds to form a hairpin structure (C). The double-stranded stem of the pri-miRNAs is recognized by the dsRNA binding protein HYL1 and the DCL1-containing processing complex (C). The association of the processing complex to the nascent primiRNA triggers processing and the subsequent production of mature miRNAs that are loaded into AGO1 (D). In some loci, and specifically with ABA treatment, HYL1 acts as a scaffold to recruit HOS15 and HDA9 to MIRNA loci (E). This interaction allows the recognition of MIRNA loci by the HOS15-HDA9 chromatin remodeling complex, which in turn will change the histone acetylation profile (E). This process leads to the repression of MIRNA gene transcription, taking them to a repressive state (A). Whether the HOS15-HDA9 complex could also regulate the proteostasis of HYL1 remains to be explored (E).
- 첨부파일